Researchers often look at genetic variations between individuals in a population to better understand phenotypic traits, such as fruit production, or human disease origin and incidence rate. One individual's genome can differ from that of the general population in numerous ways, including single base changes (single nucleotide polymorphisms, or SNPs), insertions, deletions, or even the number of gene copies. These unique differences can be used as markers in linkage and association studies that attempt to determine genes responsible for disease, plant drought tolerance, etc.
Genotyping is the process of determining the DNA sequence—the genotype—at specific positions within a gene of an individual. Genotyping can be performed by end-point or real-time PCR, sequencing, bead based hybridization detection or microarray analysis, or even mass spectrometry.
And like many technical processes, this application includes its own vocabulary. Here we provide an introduction to some of the most commonly used terms, and use them in context to draw distinctions when encountered in genotyping experiments.
Allele vs Haplotype vs Locus
| Allele | The DNA sequence at a specific chromosomal location, which presents as a variant, or alternative form, of a gene. Any given gene can have multiple different alleles. Humans have 2 sets of each chromosome so they possess the potential for only 2 alleles at any given locus, one inherited from each parent. Some genes have only one allele, such as those on the human male's Y chromosome, and any deviation from that allele can be harmful, or even fatal, to the organism. |
| Polyallelic | The existence of multiple alleles at a specific genetic locus. |
| Biallelic/Triallelic/Quatra-allelic | The number of distinct nucleotides (2/3/4) known to exist at a particular base position of an allele in a population of that species. For example, the occurrence of only A or G is a biallelic position, A or C or T is a triallelic position, and A or C or G or T is a quatra-allelic position (Figure 1). |
| Locus | A specific chromosomal location. Can refer to a gene location on a chromosome or to a specific sequence element. |
| Haplotype | A haplotype is a set of DNA variations (polymorphisms such as SNPs and indels) adjacent to one another at the same locus that tend to be inherited together (Figure 1). This set of alleles is often referred to as linked polymorphisms. |

Zygosity
| Zygosity | Describes the similarity or differences between an individual's alleles. Since most eukaryotes have 2 matching sets of chromosomes, zygosity terminology describes whether both copies of an allele, or allele-encoding trait, are the same or not. |
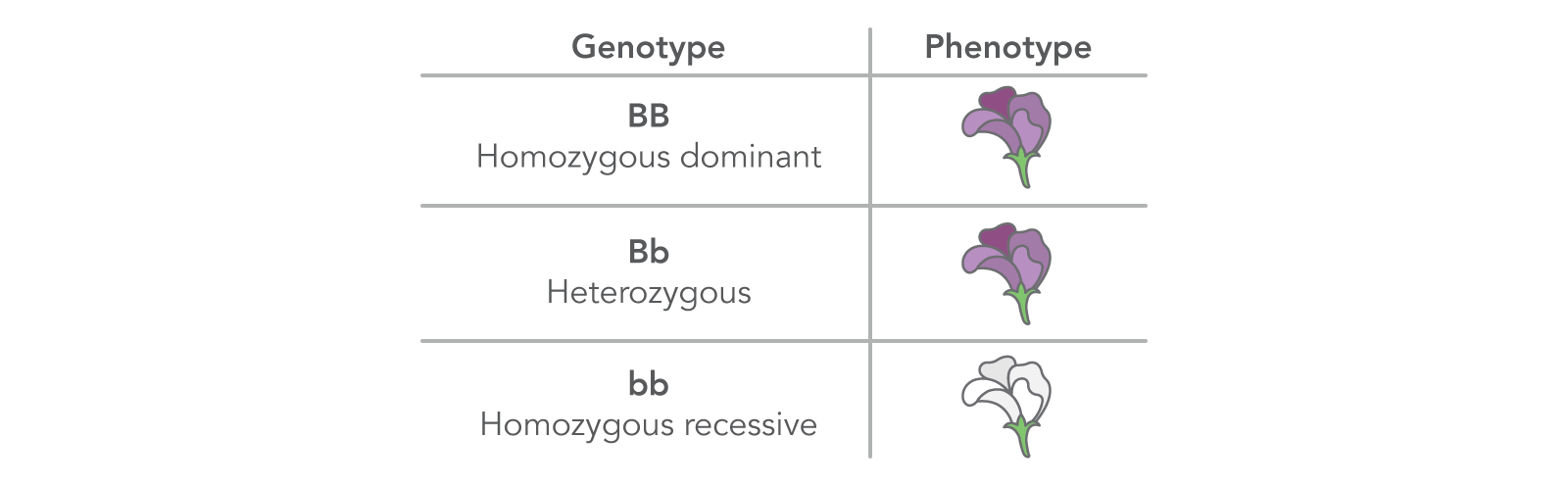
| Dominant allele (B) | A dominant allele, designated by an uppercase letter (such as "B"), always displays the phenotype it encodes. It does this either through its presence in both gene copies (BB) or by masking the expression of a second, distinct recessive allele at the same locus (Bb) (Figures 2, 3; Note: There are occasions when a recessive allele can contribute to a phenotype through co-dominance or incomplete dominance.) |
| Recessive allele (b) | A recessive allele, designated by a lowercase letter (such as "b"), expresses its associated phenotype only when paired with another recessive allele (Figures 2,3; see note under Dominant allele). |
| Homozygous (BB, bb) | An individual with 2 copies of the same allele, whether dominant (designated by 2 uppercase letters, such as "BB") or recessive (designated by 2 lowercase letters, such as "bb"). |
| Heterozygous (Bb) | An individual who has 2 different alleles for the same trait, with one dominant over the other recessive allele. |
| Hemizygous (B, b) | An individual possessing only a single copy of a gene instead of the customary 2 copies, therefore having only 1 allele. For example, all the genes on the single X and Y chromosomes in human males are hemizygous. |
| Nullzygous (B, -) |
An individual carrying two mutant alleles for the same gene, with both alleles being "null" or complete loss-of-function alleles. Can also have a complete deletion of the allele on both chromosomes. |

Allele vs Genotype
| Allele | The DNA sequence at a specific chromosomal location, which presents as a variant, or alternative form. Any given gene can have multiple different alleles. Humans have 2 sets of each chromosome so they possess the potential for only 2 alleles at any given locus, one inherited from each parent. Some genes have only one allele, such as those on the human male's Y chromosome, and any deviation from that allele can be harmful, or even fatal, to the organism. |
| Genotype | Refers broadly to the genetic makeup of an organism—its complete set of genes. Sometimes used in a narrower definition, (as in this article), genotype refers to the specific alleles found on each chromosome. |
Allele vs Phenotype
| Allelle | The DNA sequence at a specific chromosomal location, which presents as a variant, or alternative form, of a gene. Any given gene can have multiple different alleles. Humans have 2 sets of each chromosome so they possess the potential for only 2 alleles at any given locus, one inherited from each parent. Some genes have only one allele, such as those on the human male's Y chromosome, and any deviation from that allele can be harmful, or even fatal, to the organism. |
| Phenotype | The physical/observed traits determined or "expressed" by a given genotype; for example, the purple or white petals of a pea flower seen in Figure 3. |
Genotype vs Phenotype
| Genotype | Refers broadly to the genetic makeup of an organism—its complete set of genes. Sometimes used in a narrower definition, (as in this article), genotype refers to the specific alleles found on each chromosome. |
| Phenotype | The physical/observed traits determined or "expressed" by a given genotype; for example, the purple or white petals of a pea flower seen in Figure 3. |
Haplotype vs Genotype
| Haplotype | A Haplotype is a set of DNA variations (polymorphisms such as SNPs and indels) adjacent to one another at the same locus that tend to be inherited together. This set of alleles is often referred to as linked polymorphisms. |
| Genotype | Refers broadly to the genetic makeup of an organism—its complete set of genes. Sometimes used in a narrower definition, (as in this article), genotype refers to the specific alleles found on each chromosome. |
Allele vs Gene
| Allele | The DNA sequence at a specific chromosomal location, which presents as a variant, or alternative form. Any given gene can have multiple different alleles. Humans have 2 sets of each chromosome so they possess the potential for only 2 alleles at any given locus, one inherited from each parent. Some genes have only one allele, such as those on the human male's Y chromosome, and any deviation from that allele can be harmful, or even fatal, to the organism. |
| Gene | Genes are segments of DNA or RNA on a chromosome that is the functional unit of inheritance controlling the transmission and expression of traits by directing the synthesis of proteins. |
SNPs, polymorphisms, mutations, and CNVs
In human beings, 99.9% of all DNA bases in the genome—from individual to individual—are the same. The remaining 0.1% make a person unique. Each of us differs by about 10,000 non-synonymous variants from the human genome reference sequence. Of these, each of us carry around 340–400 variations that result in loss of function of certain genes [1].
An individual's genome may differ from others in numerous ways, including base differences known as single nucleotide polymorphisms (SNPs), insertions or deletions (INDELs), or differences in the number of copies of a sequence or gene (copy number variations (CNV)) (Table 1).
These variants can be:
- Harmless—Variations that cause no change in phenotype; this is true of most SNPs.
- Harmful—Variations that cause diseases, such as diabetes, cancer, heart disease, or hemophilia.
- Latent—Variations, found in coding and regulatory regions of the genome that are not harmful on their own. Their change in sequence only becomes apparent upon some type of genetic activation event.
| Reference sequence | The standard sequence for a given organism's genome, cataloged in the RefSeq database curated by the NCBI. |
| Polymorphism | Variation at a genomic locus carried by a percentage of individuals within a population, thus creating different genotypes across that population. |
| Single-nucleotide polymorphism (SNP) | Variation in a single nucleotide that occurs at a specific position in the genome. To be considered a SNP, the variation must be present in >1% of the population. Less than this, and it would be considered a rare variant (abnormal change). |
| Single nucleotide variation (SNV) | A base variation, distinct from the reference sequence, without information regarding how often this variation occurs. |
| Multiple nucleotide polymorphism (MNP) | When 2 or more SNPs occur right next to each other in the same haplotype, see Table 1. |
| INDEL (INsertion/DELetion) |
Sequence that has been inserted or deleted in one genome relative to another. A deletion in one genome corresponds to an insertion in the other. |
| Mutation | Changes in DNA sequence from an individual's inherited genetic sequence (as conferred in the reference sequence for that individual). Each of the above types of polymorphisms—SNPS, SNVs, MNPs, INDELs—are considered mutations. However, while polymorphisms are defined as being present within an appreciable subset of the general population, mutations also include alterations in DNA sequence that are rare or have been identified in just a single individual. |
| Copy number variation (CNV) | When the number of copies of a particular genetic sequence differs between individuals. It is caused by repeats in the genome, the number of which can vary dramatically across a population. |
| Type | Reference sequence | Alternate sequence |
|---|---|---|
| SNP (single) | T | G |
| MNP (multiple) | TA | GC |
| Insertion | AGT | ACGT |
| ATCGGG | ATCTGAGGG | |
| Deletion | ACGT | AGT |
| ATCTGAGGG | ATCGGG |
Table 1. Distinction between SNPs vs. MNPs vs. InDELs.
Types of mutations
| Germline mutation | A mutation present in one's gametes (egg or sperm), and thus, can be inherited. Germline mutations are responsible for familial inherited diseases, such as retinoblastoma, Huntington's disease, and cystic fibrosis. They can be either dominant or recessive mutations, requiring only 1 or both alleles, respectively, to be mutated for expression of the inherited trait. |
| Somatic mutation | A mutation that occurs in non-germline tissues and cannot be inherited. Thus, such mutations are only present in some of the cells of the body (e.g., in a tumor), giving rise to the presence of multiple genotypes within a single individual. |
| Silent mutations | A mutation that does not have a visible/detectable effect on the phenotype of an organism. |
| Non-synonymous variant | A SNP that changes the codon it resides in, resulting in an altered amino acid sequence for the encoded protein (missense mutation) or a truncated protein (nonsense mutation). |
| Missense mutation | A mutation at a single base that results in the encoding of a distinct amino acid in the resulting protein. The amino acid substitution may render the protein fully functional, partially functional, or nonfunctional. |
| Nonsense mutation | A mutation that results in a codon change to a chain-terminating codon, thus generating a truncated protein. Such proteins are often nonfunctional. |
Allele frequency
| Minor allele frequency (MAF) | The frequency (percent or fraction) of the second most common allele for a given locus in a population. |
| MAF/MinorAlleleCount (Figure 4) | An equation that provides an estimate of the number of times a particular SNP has been observed in the population used in a specific study. For example:
C=0.1506/754(1000 Genomes)
where, C = the minor allele for that particular locus 0.1506 = the frequency of the C allele (MAF); in this case meaning 15% within the 1000 Genomes database 754 = the number of times this SNP has been observed in the population of the study |
The SNP database, dbSNP, is the NCBI-curated database of verified SNPs, each with its own RefSNP entry. Figure 4 shows a RefSNP entry for a pathogenic SNV, noting the alleles detected and the MAF/MinorAlleleCount of these alleles, based on reference sequences from 4 different sources.

Your genotyping resource
Reagents for genotyping. IDT offers multiple genotyping solutions, the rhAmp™ SNP Genotyping System, locked nucleic acid qPCR probes called Affinity Plus qPCR Probes, MGB Eclipse® Probes that help to enable the identification of small sequence alterations.
Learn more about these IDT genotyping solutions.
A complete PCR-based SNP genotyping solution. The rhAmp SNP Genotyping System includes a predesigned assay collection addressing >10 million human SNPs, including a broad selection of functionally substantiated absorption, distribution, metabolism, and excretion (ADME) SNP assays. A custom assay design pipeline is also available for newly discovered human SNPs or assay designs of other species. The design of rhAmp SNP assays makes it possible to identify SNPs in difficult sequence regions with amplicon lengths as short as 40 bp.
Learn more about the rhAmp SNP Genotyping System.
Technical support. In addition to the comprehensive set of tools, reagents, and educational resources for PCR-based SNP genotyping, IDT also provides world-class technical support. These scientists are available to answer all types of genotyping questions ranging from experimental design to interpreting data. Contact us with your questions about genotyping assay design at applicationsupport@idtdna.com.