When CRISPR genome editing was introduced in 2009, plasmids were generously made available by the labs where they were developed so that other researchers around the world could use them to express Cas proteins in CRISPR experiments. These plasmids have been cited in numerous publications and have helped to advance understanding and applications of CRISPR-Cas genome editing. Meanwhile, however, many discoveries have been made about the best approaches to CRISPR genome editing—and it turns out that CRISPR plasmids are often not the most efficient approach to use, due to undesired effects such as cytotoxicity and unpredictability, both of which are discussed below.
Why use ribonucleoprotein?
Some cell types cannot tolerate plasmid transfection, so one important consideration is direct cytotoxicity due to plasmid transfection itself. Indeed, research shows that transfecting some cell types with any plasmid may cause cell death [1], while some transfection reagents (e.g., lipids) used for plasmid transfection are themselves toxic to cells [2]. Importantly, it has also been shown that plasmid transfections, specifically in CRISPR experiments, can cause cytotoxicity in embryonic stem cells [3], which could be of concern to many researchers using this cell type.
Three ways plasmids can complicate CRISPR genome editing
Even with cells that can tolerate plasmid transfections, there are other problems associated with the use of plasmids for CRISPR editing, such as complications in the timing of CRISPR experiments.
- The most obvious disadvantage in timing is that preparing plasmids is extra work, which wastes valuable bench time (see the infographic).
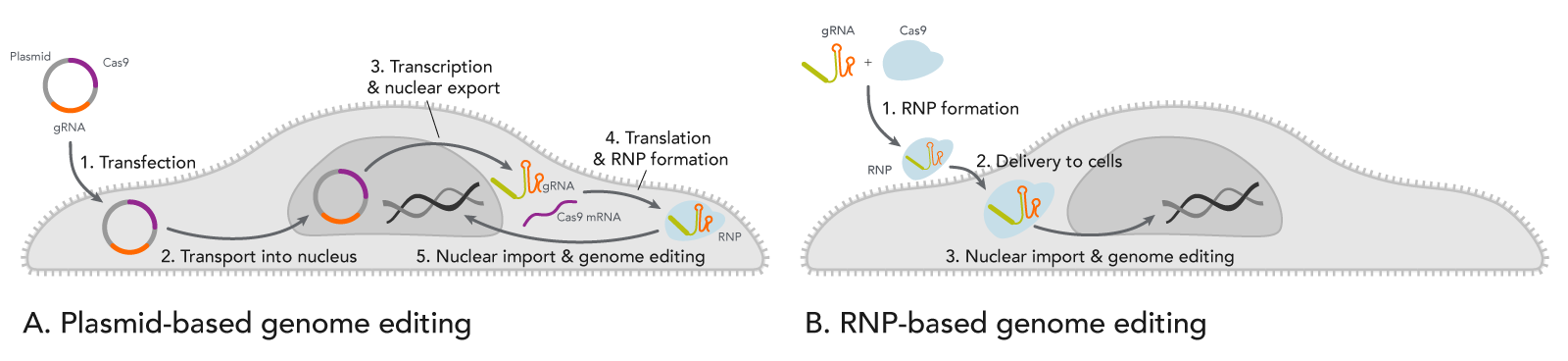
- Transfecting a plasmid creates a waiting time before the CRISPR genome editing can begin. As shown in Figure 1A, this is a complex, multi-step process that may require around 24 hours. The uncertainty of when the enzyme is expressed may be problematic for experiments that require other manipulations such as timed drug delivery or delivery of a donor template for homology-directed recombination (HDR).
![20_GE_CRISPR-RNP-vs-Plasmid-cells CRISPR plasmid transfection slows genome editing compared to direct RNP introduction.]() Figure 1. Comparison of events after introducing CRISPR plasmid vs. ribonucleoprotein (RNP) into cells. (A) When cells are transfected with a CRISPR plasmid, the plasmid is transported into the nucleus for RNA transcription of both gRNA and Cas-encoding mRNA. Next, the resulting mRNA for the Cas enzyme is exported from the nucleus where it is translated into protein. The nascent protein is then folded. Meanwhile, the gRNA is also transcribed from the plasmid(s) and exported from the nucleus. The Cas enzyme and the gRNA bind to form a ribonucleoprotein (RNP) and are transported back into the nucleus where genome editing occurs. (B) When pre-formed RNP is introduced by nucleofection into cells, the enzyme rapidly starts cutting targeted genomic DNA.
Figure 1. Comparison of events after introducing CRISPR plasmid vs. ribonucleoprotein (RNP) into cells. (A) When cells are transfected with a CRISPR plasmid, the plasmid is transported into the nucleus for RNA transcription of both gRNA and Cas-encoding mRNA. Next, the resulting mRNA for the Cas enzyme is exported from the nucleus where it is translated into protein. The nascent protein is then folded. Meanwhile, the gRNA is also transcribed from the plasmid(s) and exported from the nucleus. The Cas enzyme and the gRNA bind to form a ribonucleoprotein (RNP) and are transported back into the nucleus where genome editing occurs. (B) When pre-formed RNP is introduced by nucleofection into cells, the enzyme rapidly starts cutting targeted genomic DNA. - Another timing issue caused by plasmids is that Cas enzyme and gRNA can remain active in the cells for prolonged periods. CRISPR genome editing experiments have a propensity for off-target effects (OTEs). OTEs are undesired changes to genomic DNA sequences with some sequence similarity to the target site. OTEs occur when the Cas enzyme is directed to the wrong site by the gRNA due to such sequence similarity. With good gRNA design, the chances of OTEs occurring are relatively low compared to the chance of the Cas enzyme being guided to the target site. However, if the Cas enzyme and the gRNA are present in the cell for an extended period of time, the chance of OTEs is increased [2]. With plasmid transfections, the cell continues expressing the Cas enzyme and the gRNA over several days [3]. The longer the Cas protein and gRNA persist, the greater the chance that genomic DNA in non-targeted sites will be cut [4] (see the infographic).
Significant OTEs may themselves cause cytotoxicity even if the plasmid does not. OTEs may also confound the experimental data. Furthermore, in certain applications, OTEs may cause harm to the organism. For example, an OTE in an oncogenic region could cause cancer [3]. Hence, Cas enzyme should not be expressed for longer than necessary.
Plasmids give you unpredictable variability
Even if OTEs are of no major concern in a specific experiment, plasmid transfections will result in variable editing efficiencies because of the uncontrollability of gRNA and Cas enzyme expression levels (see the infographic). Other unpredictable and unwanted events are possible when using plasmids. For example, even though plasmid transfection is ostensibly “transient,” a plasmid (or a part of it) can become incorporated into the cell genome, forming a “footprint” that could confound experimental results (see the infographic).
What about stable expression, mRNA, or viral delivery of CRISPR components?
Different Cas enzymes have been expressed by stable integration in mammalian cell lines. This leads to constitutive expression of these nucleases. While interesting, this does not replicate a natural state, so relevancy can be called into question. Furthermore, the issue of long-term Cas expression leading to OTEs, which is a problem with plasmids as described above, also exists with stable expression of the Cas enzyme. Importantly, Cas9 gene integration into the genome of mammalian cells has been reported in experiments when stable integration was not intended [5]. This is evidence that plasmids (and other vectors such as viruses) carrying any Cas gene are too dangerous for direct use in humans [5].
Transfection of mRNA encoding a Cas enzyme is more efficient than plasmid transfection, because using mRNAs saves time by eliminating the transcription step. However, mRNA transfection still relies on the cell to translate the mRNA and fold the nascent Cas protein, resulting in variability in timing. An additional disadvantage of transfecting mRNAs is that they are prone to degradation by RNases. Handling mRNA long enough to express a large protein such as a Cas enzyme can be challenging even in a very clean environment.
Using viral vectors for delivery of CRISPR components requires more work than transfecting plasmids, because there are many time- and labor-intensive steps necessary to produce viral vectors. Viral vectors are also more expensive to produce than plasmids. In addition, with viral vectors, as with plasmids, there remains unpredictability in timing while waiting for the cells to produce Cas and gRNA. Using viral vectors also introduces the risk of incorporating viral components or a Cas gene into the target genome, which may confound experimental results or produce situations hazardous to human health [5]. Furthermore, viral components themselves (e.g., proteins and glycoproteins) may interfere with experimental results and cause immune responses.
What is the best alternative to using plasmids for CRISPR?
Recombinant Cas enzymes and chemically synthesized gRNA are the recommended reagents for CRISPR genome editing. For increased efficiency, the Cas enzyme and the gRNA can be combined on the bench before being delivered into the cells. The Cas-gRNA complex is known as a ribonucleoprotein (RNP). A number of methods have been developed for direct delivery of RNPs to cells. Frequently, RNP is delivered into cells in culture by lipofection or electroporation (Figure 1B). Electroporation using a nucleofection protocol is often preferred, as this allows the RNP to quickly enter the nucleus of cells and start cutting the genome.
The RNP complex is active immediately because it is already fully developed. Equally importantly, the natural cellular mechanisms of protein and RNA turnover ensure that the RNP will be destroyed. In the absence of plasmid, the RNP is not replaced, greatly decreasing the chances of OTEs being introduced. Without a plasmid or Cas-encoding gene, there is no risk of a molecular footprint being introduced.
Can all CRISPR-Cas systems be delivered in RNP format?
Not all publicly available Cas enzymes work well in RNP format. As shown by IDT scientists, some high-precision Cas enzymes which may have reasonable DNA-cutting efficiency when expressed from plasmids do not function well when used in RNP format [6]. Thus, it is essential to use a Cas enzyme that is known to work well in RNP format. Alt‑R™ Cas9 V3, HiFi Cas9, and Cas12a Ultra nucleases all meet this requirement.
RNPs for CRISPR in animal models
In a relatively simple animal model such as zebrafish, injection of eggs with RNP is highly successful, as shown by University of Utah scientists in collaboration with IDT [7]. Furthermore, in adult mammals, recent studies have shown that RNP delivery in vivo is possible using nanoparticles. Gold nanoparticles [8] are popular, although other types of nanoparticles (e.g., zinc) are also under investigation [9]. Nanoparticles have been shown to be very efficient at delivering RNPs into animals [2,10,11]. As mentioned above, avoiding OTEs is extremely important in certain CRISPR applications, so it is recommended to avoid using CRISPR plasmids.
RNPs improve enzyme and guide RNA efficiency
There are additional possible advantages available when using RNPs instead of plasmids for CRISPR editing. Not only are high-fidelity recombinant Cas enzymes (Alt‑R HiFi Cas9 and Cas12a Ultra) available, but additionally, the Alt‑R guide RNAs for Cas9 or Cas12a are chemically modified to enhance editing efficiency and to decrease toxicity. IDT chemical modifications also decrease the risk of an immune response to the gRNA in mammalian systems. Combining optimized Alt‑R guide RNA with an Alt‑R Cas enzyme speeds up CRISPR genome editing projects and delivers reliable results.